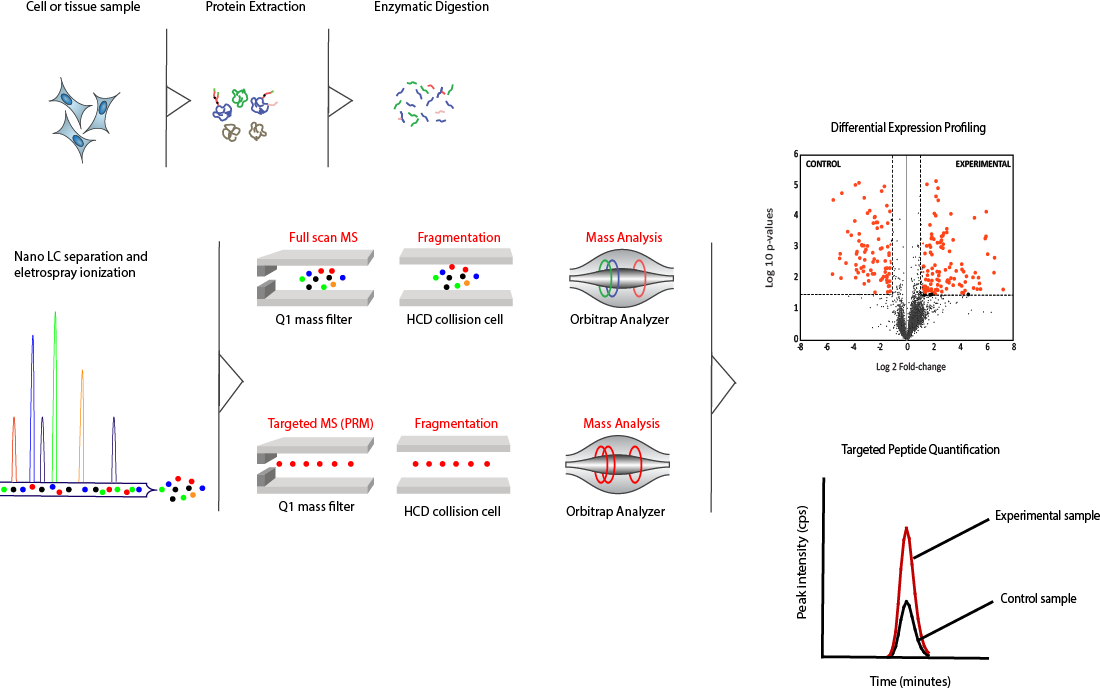
Technology Overview using High Resolution Mass Spectrometry (HRMS)
Our technological approach for protein identification and quantitation can be divided into two strategies : a global, discovery-based approach, and a more targeted, hypothesis-driven approach.
Proteins from tissues or cell culture are first extracted then enzymatically digested to generate a complex mixture of peptides. This mixture is first separated chromatographically using nanoscale capillary columns, then transferred to the gaseous phase by electrospray ionization.
In the discovery-based detection strategy, all peptide ions are detected and sequenced by the Orbitrap mass analyzer. This provides us with a global profile of all the components present in our peptide mixture using full scan LC-MS. To focus on a smaller subset of proteins of interest, an alternative detection strategy (parallel reaction monitoring, or PRM) utilizing a target list of peptides is applied
Peptides can then be identified and quantified from the intensities of their corresponding mass spectra. The diagram below summarizes the overall workflow used for protein identification and quantitation. Our proteomics driven approaches enable the parallel multiplexing of protein detection and quantification with a high degree of accuracy and confidence.
Overall Workflow Diagram